[Past Projects]
Dr. Rachel Olson
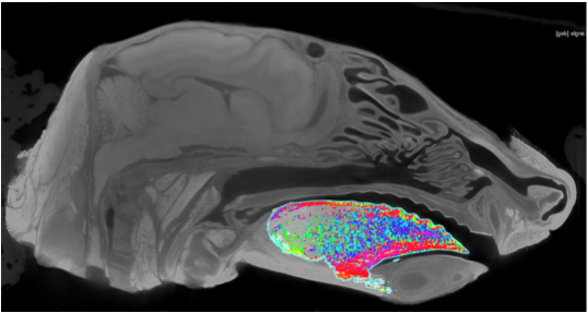
Primary Goals
- Digitally dissect the muscles involved in chewing (multiple individuals and species have been scanned).
- Determine the fiber orientations of the tongue.
- Potential to develop an analysis method to compare different species.
- Potential to apply anatomical data of the tongue to in vivo biomechanics of the tongue.
- Anatomical imaging – learn about and look at CT scans and x-ray imaging. There may be opportunities to participate in collecting this data.
- Digital dissection methods using image processing software like Slicermorph and VGstudio.
- Interact with and learn anatomy and biomechanics in an applied context.
- Contrast-enhanced staining methods in the wet lab.
- Possibility of traditional gross dissections.
- Data visualization and statistical analysis in R.
- Participate in lab meetings and scientific discussions.
- Participate in the Biological Undergraduate Research Symposium.
Click here for more information about Dr. Olson’s lab
