[Past Projects]
Dr. Steve Weeks and Chathumadavi Ediriweera

Why clam shrimps?
Most of the animals, especially including mammals and humans, are at an advanced stages of chromosomal evolution. Therefore, it is difficult to study sex chromosome evolution in these species. Clam shrimps, on the other hand, are in an early stage of sex chromosome evolution. This allows us to see into the stages of sex chromosome evolution at a stage when theory suggests such chromosomes will begin to degrade. Clam shrimps are androdioecious (males + hermaphrodites) and have a WZ sex-determining system which prevents major chromosome degradation by expressing both sex chromosomes in homozygous form (WW and ZZ). This keeps the W chromosome from degrading. Hence, Clam shrimp are an excellent model system to investigate how advanced degradation develops in other species, such as humans.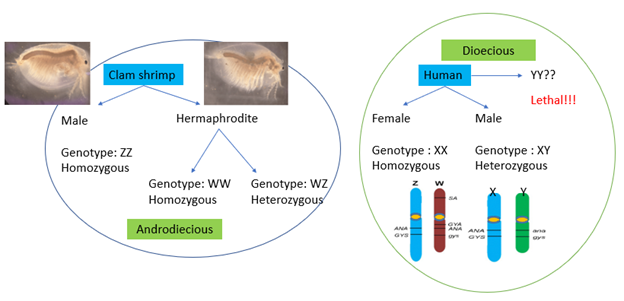
Project details:
We have a whole genome sequence of clam shrimp. The genome is like a book and the chromosomes are its chapters. We are specifically interested in one chapter: the sex chromosome. We are using bioinformatic pipelines to analyze genomic data specifically related to sex chromosomes in order to test hypotheses of sex chromosomal evolution. Sex chromosome evolutionary theory predicts that a high proportion of repeat sequences/Transposable Elements (TEs) should accumulate on incipient sex chromosomes. An important next step would be to annotate this genome content in terms of repeat sequences. The proposed clam shrimp genome-wide TE analysis will: 1) identify novel TEs and enhance the genome-wide content annotation; 2) improve our understanding of the processes of sex chromosome degradation in E. texana; and 3) substantially broaden our understanding of sex chromosome evolution in animals overall.
Benefits for the student:
- Learn how to access and utilize databases such as NCBI, Blast, ClustalW, and Structure.
- Understand how sequence homology relates to evolution.
- Learn how to manage and automate big data using a unix platform and python programming language.
- Learn how to analyze and visualize data using R programming language.
- Understand the power of Bioinformatics.
Qualifications:
We seek undergraduate students who are interested in bioinformatics to study sex chromosome evolution from any major who is willing to learn the above skills.Click here for more information on the Weeks lab
